PUBLICATIONS
Vaccination with an HIV T- cell immunogen induces alterations in the mouse gut microbiota
Authors: Alessandra Borgognone; Aleix Elizalde- Torrent; Maria Casadellà; Luis Romero; Tuixent Escribà; Mariona Parera; Francesc Català-Moll; Marc Noguera- Julian; Christian Brander; Alex Olvera; Roger Paredes
ABSTRACT:
The gut microbiota is emerging as a crucial factor modulating vaccine responses; however, few studies have investigated if vaccines, in turn, can alter the microbiota and to what extent such changes may improve vaccine efficacy. To understand the effect of T-cell vaccination on the gut microbiome, we administered an HIV-1 T-cell immunogen (HTI arm) or PBS (control, Mock arm) to C57Bl/6 mice following a heterologous prime-boost scheme. The longitudinal dynamics of the mice gut microbiota was characterized by 16 S ribosomal RNA sequencing in fecal samples collected from cages, as well as from three gut sections (cecum, small and large intestine). Serum and spleen cells were obtained at the last time point of the study to assess immune correlates using IFNγ ELISPOT and cytokine Luminex® assays. Compared with Mock, HTI-vaccinated mice were enriched in Clostridiales genera (Eubacterium xylanophilum group, Roseburia and Ruminococcus) known as primary contributors of anti-inflammatory metabolites, such as short-chain fatty acids. Such shift was observed after the first HTI dose and remained throughout the study follow-up (18 weeks). However, the enriched Clostridiales genera were different between feces and gut sections. The abundance of bacteria enriched in vaccinated animals positively correlated with HTI-specific T-cell responses and a set of pro-inflammatory cytokines, such as IL-6. This longitudinal analysis indicates that, in mice, T-cell vaccination may promote an increase in gut bacteria known to produce anti-inflammatory molecules, which in turn correlate with proinflammatory cytokines, suggesting an adaptation of the gut microbial milieu to T-cell-induced systemic inflammation.
DOI: https://doi.org/10.1038/s41522-022-00368-y
JOURNAL: NPJ Biofilms and Microbiomes
DATE: 2022 Dec 30
Probiotic effects on immunity and microbiome in HIV-1 discordant patients
Authors: Carlos Blázquez- Bondia; Mariona Parera; Francesc Català-Moll; Maria Casadellà; Aleix Elizalde- Torrent; Meritxell Aguiló; Jordi Espadaler- Mazo; José Ramon Santos; Roger Paredes; Marc Noguera-Julian
ABSTRACT:
Background: Some HIV-1 infected patients are unable to completely recover normal CD4+ T-cell (CD4+) counts after achieving HIV-1 suppression with combined Antiretroviral Therapy (cART), hence being classified as immuno-discordant. The human microbiome plays a crucial role in maintaining immune homeostasis and is a potential target towards immune reconstitution.
Setting: RECOVER (NCT03542786) was a double-blind placebo-controlled clinical trial designed to evaluate if the novel probiotic i3.1 (AB-Biotics, Sant Cugat del Vallès, Spain) was able to improve immune reconstitution in HIV-1 infected immuno-discordant patients with stable cART and CD4+ counts <500 cells/mm3. The mixture consisted of two strains of L. plantarum and one of P. acidilactici, given with or without a fiber-based prebiotic.
Methods: 71 patients were randomized 1:2:2 to Placebo, Probiotic or probiotic + prebiotic (Synbiotic), and were followed over 6 months + 3-month washout period, in which changes on systemic immune status and gut microbiome were evaluated. Primary endpoints were safety and tolerability of the investigational product. Secondary endpoints were changes on CD4+ and CD8+ T-cell (CD8+) counts, inflammation markers and faecal microbiome structure, defined by alpha diversity (Gene Richness), beta diversity (Bray-Curtis) and functional profile. Comparisons across/within groups were performed using standard/paired Wilcoxon test, respectively.
Results: Adverse event (AE) incidence was similar among groups (53%, 33%, and 55% in the Placebo, Probiotic and Synbiotic groups, respectively, the most common being grade 1 digestive AEs: flatulence, bloating and diarrhoea. Two grade 3 AEs were reported, all in the Synbiotic group: abdominal distension (possibly related) and malignant lung neoplasm (unrelated), and 1 grade 4 AE in the Placebo: hepatocarcinoma (unrelated). Synbiotic exposure was associated with a higher increase in CD4+/CD8+ T-cell (CD4/CD8) ratio at 6 months vs baseline (median=0.76(IQR=0.51) vs 0.72(0. 45), median change= 0.04(IQR=0.19), p = 0.03). At month 9, the Synbiotic group had a significant increase in CD4/CD8 ratio (0.827(0.55) vs 0.825(0.53), median change = 0.04(IQR=0.15), p= 0.02) relative to baseline, and higher CD4+ counts (447 (157) vs. 342(73) counts/ml, p = 0.03), and lower sCD14 values (2.16(0.67) vs 3.18(0.8), p = 0.008) than Placebo. No effect in immune parameters was observed in the Probiotic arm. None of the two interventions modified microbial gene richness (alpha diversity). However, intervention as categorical variable was associated with slight but significant effect on Bray-Curtis distance variance (Adonis R2 = 0.02, p = 0.005). Additionally, at month 6, Synbiotic intervention was associated with lower pathway abundances vs Placebo of Assimilatory Sulphate Reduction (8.79·10-6 (1.25·10-5) vs. 1.61·10-5 (2.77·10-5), p = 0.03) and biosynthesis of methionine (2.3·10-5 (3.17·10-5) vs. 4·10-5 (5.66·10-5), p = 0.03) and cysteine (1.83·10-5 (2.56·10-5) vs. 3.3·10-5 (4.62·10-5), p = 0.03). At month 6, probiotic detection in faeces was associated with significant decreases in C Reactive Protein (CRP) vs baseline (11.1(22) vs. 19.2(66), median change= -2.7 (13.2) ug/ml, p = 0.04) and lower IL-6 values (0.58(1.13) vs. 1.17(1.59) ug/ml, p = 0.02) when compared with samples with no detectable probiotic. No detection of the probiotic was associated with higher CD4/CD8 ratio at month 6 vs baseline (0.718(0.57) vs. 0.58(0.4), median change = 0.4(0.2), p = 0.02). After washout, probiotic non-detection was also associated with a significant increase in CD4+ counts (457(153) vs. 416(142), median change = 45(75), counts/ml, p = 0.005) and CD4/CD8 ratio (0.67(0.5) vs 0.59(0.49), median change = 0.04 (0.18), p = 0.02).
Conclusion: A synbiotic intervention with L. plantarum and P. acidilactici was safe and led to small increases in CD4/CD8 ratio and minor reductions in sCD14 of uncertain clinical significance. A probiotic with the same composition was also safe but did not achieve any impact on immune parameters or faecal microbiome composition.
Gut microbiome signatures linked to HIV-1 reservoir size and viremia control
Authors: Alessandra Borgognone; Bruna Oriol- Tordera; Beatriz Mothe; Roger Paredes; Maria C. Puertas
ABSTRACT:
Background
The potential role of the gut microbiome as a predictor of immune-mediated HIV-1 control in the absence of antiretroviral therapy (ART) is still unknown. In the BCN02 clinical trial, which combined the MVA.HIVconsv immunogen with the latency-reversing agent romidepsin in early-ART treated HIV-1 infected individuals, 23% (3/13) of participants showed sustained low-levels of plasma viremia during 32 weeks of a monitored ART pause (MAP). Here, we present a multi-omics analysis to identify compositional and functional gut microbiome patterns associated with HIV-1 control in the BCN02 trial.
Results
Viremic controllers during the MAP (controllers) exhibited higher Bacteroidales/Clostridiales ratio and lower microbial gene richness before vaccination and throughout the study intervention when compared to non-controllers. Longitudinal assessment indicated that the gut microbiome of controllers was enriched in pro-inflammatory bacteria and depleted in butyrate-producing bacteria and methanogenic archaea. Functional profiling also showed that metabolic pathways related to fatty acid and lipid biosynthesis were significantly increased in controllers. Fecal metaproteome analyses confirmed that baseline functional differences were mainly driven by Clostridiales. Participants with high baseline Bacteroidales/Clostridiales ratio had increased pre-existing immune activation-related transcripts. The Bacteroidales/Clostridiales ratio as well as host immune-activation signatures inversely correlated with HIV-1 reservoir size.
Conclusions
The present proof-of-concept study suggests the Bacteroidales/Clostridiales ratio as a novel gut microbiome signature associated with HIV-1 reservoir size and immune-mediated viral control after ART interruption.
PREVIOUS RELATED PUBLICATIONS
Associations of the gut microbiome and clinical factors with acute GVHD in allogeneic HSCT recipients
AUTHORS: Ilett EE, Jørgensen M, Noguera-Julian M, Nørgaard JC, Daugaard G, Helleberg M, Paredes R, Murray DD, Lundgren J, MacPherson C, Reekie J, Sengeløv H.
ABSTRACT:
Acute graft-versus-host disease (aGVHD) is a leading cause of transplantation-related mortality after allogeneic hematopoietic stem cell transplantation (aHSCT). 16S ribosomal RNA (16S rRNA) gene-based studies have reported that lower gut bacterial diversity and the relative abundance of certain bacteria after aHSCT are associated with aGVHD. Using shotgun metagenomic sequencing and a large cohort, we aimed to confirm and extend these observations. Adult aHSCT recipients with stool samples collected from day -30 to day 100 relative to aHSCT were included. One sample was selected per patient per period (pre-aHSCT (day -30 to day 0), early post-aHSCT (day 1 to day 28), and late post-aHSCT (day 29 to day 100)), resulting in 150 aHSCT recipients and 259 samples. Microbial and clinical factors were tested for differences between time periods and an association with subsequent aGVHD. Patients showed a decline in gut bacterial diversity posttransplant, with several patients developing a dominance of Enterococcus. A total of 36 recipients developed aGVHD at a median of 34 days (interquartile range, 26-50 days) post-aHSCT. Lower microbial gene richness (P = .02), a lower abundance of the genus Blautia (P = .05), and a lower abundance of Akkermansia muciniphila (P = .01) early post-aHSCT was observed in those who developed aGVHD. Myeloablative conditioning was associated with aGVHD along with a reduction in gene richness and abundance of Blautia and A muciniphila. These results confirm low diversity and Blautia being associated with aGVHD. Crucially, we add that pretransplant conditioning is associated with changes in gut microbiota. Investigations are warranted to determine the interplay of gut microbiota and conditioning in the development of aGVHD.
The Impact of Human Immunodeficiency Virus Infection on Gut Microbiota α-Diversity: An Individual-level Meta-analysis
AUTHORS: Tuddenham SA, Koay WLA, Zhao N, White JR, Ghanem KG, Sears CL; HIV Microbiome Re-analysis Consortium
ABSTRACT:
Background: Whether human immunodeficiency virus (HIV) infection impacts gut microbial α-diversity is controversial. We reanalyzed raw 16S ribosomal RNA (rRNA) gene sequences and metadata from published studies to examine α-diversity measures between HIV-uninfected (HIV-) and HIV-infected (HIV+) individuals.
Methods: We conducted a systematic review and individual level meta-analysis by searching Embase, Medline, and Scopus for original research studies (inception to 31 December 2017). Included studies reported 16S rRNA gene sequences of fecal samples from HIV+ patients. Raw sequence reads and metadata were obtained from public databases or from study authors. Raw reads were processed through standardized pipelines with use of a high-resolution taxonomic classifier. The χ2 test, paired t tests, and generalized linear mixed models were used to relate α-diversity measures and clinical metadata.
Results: Twenty-two studies were identified with 17 datasets available for analysis, yielding 1032 samples (311 HIV-, 721 HIV+). HIV status was associated with a decrease in measures of α-diversity (P < .001). However, in stratified analysis, HIV status was associated with decreased α-diversity only in women and in men who have sex with women (MSW) but not in men who have sex with men (MSM). In analyses limited to women and MSW, controlling for HIV status, women displayed increased α-diversity compared with MSW.
Conclusions: Our study suggests that HIV status, sexual risk category, and gender impact gut microbial community α-diversity. Future studies should consider MSM status in gut microbiome analyses.
Gut microbiome comparability of fresh-frozen versus stabilized-frozen samples from hospitalized patients using 16S rRNA gene and shotgun metagenomic sequencing
AUTHORS: Ilett EE, Jørgensen M, Noguera-Julian M, Daugaard G, Murray DD, Helleberg M, Paredes R, Lundgren J, Sengeløv H, MacPherson C.
ABSTRACT:
Collection of faecal samples for microbiome analysis in acutely sick patients is logistically difficult, particularly if immediate freezing is required (i.e. fresh-frozen, or FF sampling). Previous studies in healthy/non-hospitalized volunteers have shown that chemical stabilization (i.e. stabilized-frozen, or SF sampling) allows room-temperature storage with comparable results to FF samples. To test this in a hospital setting we compared FF and SF approaches across 17 patients undergoing haematopoietic stem cell transplantation (HSCT) using both 16S rRNA gene and shotgun metagenomic sequencing. A paired (same stool specimen) comparison of FF and SF samples was made, with an overall comparable level in relative taxonomic abundances between the two sampling techniques. Though shotgun metagenomic sequencing found significant differences for certain bacterial genera (P < 0.001), these were considered minor methodological effects. Within-sample diversity of either method was not significantly different (Shannon diversity P16SrRNA = 0.68 and Pshotgun = 0.89) and we could not reject the null hypothesis that between-sample variation in FF and SF were equivalent (P16SrRNA = 0.98 and Pshotgun = 1.0). This indicates that SF samples can be used to reliably study the microbiome in acutely sick patient populations, thus creating and enabling further outcomes-based metagenomic studies on similarly valuable cohorts.
Evolution of the gut microbiome following acute HIV-1 infection
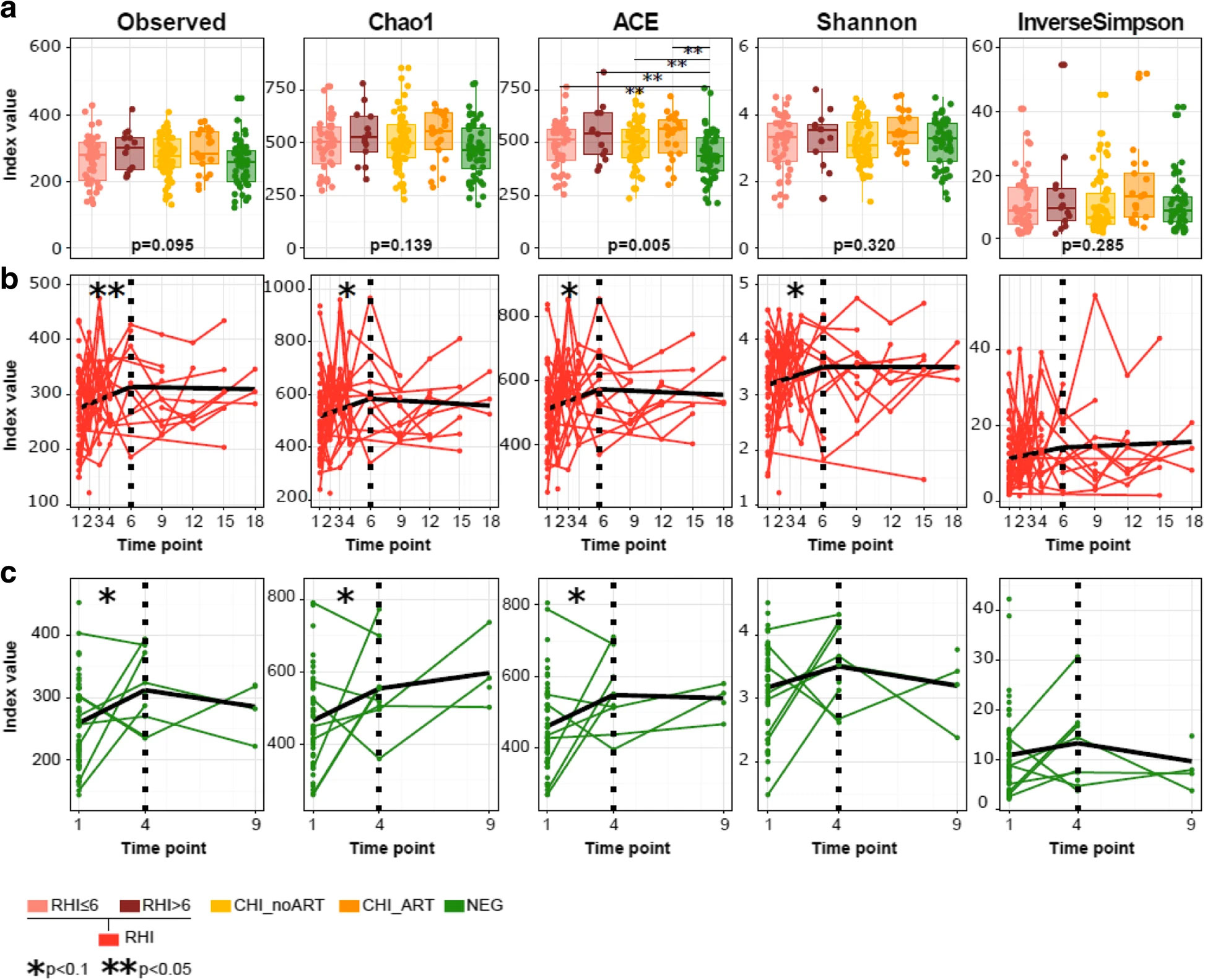
AUTHORS: Rocafort M, Noguera-Julian M, Rivera J, Pastor L, Guillén Y, Langhorst J, Parera M, Mandomando I, Carrillo J, Urrea V, Rodríguez C, Casadellà M, Calle ML, Clotet B, Blanco J, Naniche D, Paredes R.
ABSTRACT:
Background: In rhesus macaques, simian immunodeficiency virus infection is followed by expansion of enteric viruses but has a limited impact on the gut bacteriome. To understand the longitudinal effects of HIV-1 infection on the human gut microbiota, we prospectively followed 49 Mozambican subjects diagnosed with recent HIV-1 infection (RHI) and 54 HIV-1-negative controls for 9-18 months and compared them with 98 chronically HIV-1-infected subjects treated with antiretrovirals (n = 27) or not (n = 71).
Results: We show that RHI is followed by increased fecal adenovirus shedding, which persists during chronic HIV-1 infection and does not resolve with ART. Recent HIV-1 infection is also followed by transient non-HIV-specific changes in the gut bacterial richness and composition. Despite early resilience to change, an HIV-1-specific signature in the gut bacteriome-featuring depletion of Akkermansia, Anaerovibrio, Bifidobacterium, and Clostridium-previously associated with chronic inflammation, CD8+ T cell anergy, and metabolic disorders, can be eventually identified in chronically HIV-1-infected subjects.
Conclusions: Recent HIV-1 infection is associated with increased fecal shedding of eukaryotic viruses, transient loss of bacterial taxonomic richness, and long-term reductions in microbial gene richness. An HIV-1-associated microbiome signature only becomes evident in chronically HIV-1-infected subjects.
Low nadir CD4+ T-cell counts predict gut dysbiosis in HIV-1 infection
AUTHORS: Guillén Y, Noguera-Julian M, Rivera J, Casadellà M, Zevin AS, Rocafort M, Parera M, Rodríguez C, Arumí M, Carrillo J, Mothe B, Estany C, Coll J, Bravo I, Herrero C, Saz J, Sirera G, Torrella A, Navarro J, Crespo M, Negredo E, Brander C, Blanco J, Calle ML, Klatt NR, Clotet B, Paredes R.
ABSTRACT:
Human immunodeficiency virus (HIV)-1 infection causes severe gut and systemic immune damage, but its effects on the gut microbiome remain unclear. Previous shotgun metagenomic studies in HIV-negative subjects linked low-microbial gene counts (LGC) to gut dysbiosis in diseases featuring intestinal inflammation. Using a similar approach in 156 subjects with different HIV-1 phenotypes, we found a strong, independent, dose-effect association between nadir CD4+ T-cell counts and LGC. As in other diseases involving intestinal inflammation, the gut microbiomes of subjects with LGC were enriched in gram-negative Bacteroides, acetogenic bacteria and Proteobacteria, which are able to metabolize reactive oxygen and nitrogen species; and were depleted in oxygen-sensitive methanogenic archaea and sulfate-reducing bacteria. Interestingly, subjects with LGC also showed increased butyrate levels in direct fecal measurements, consistent with enrichment in Roseburia intestinalis despite reductions in other butyrate producers. The microbiomes of subjects with LGC were also enriched in bacterial virulence factors, as well as in genes associated with beta-lactam, lincosamide, tetracycline, and macrolide resistance. Thus, low nadir CD4+ T-cell counts, rather than HIV-1 serostatus per se, predict the presence of gut dysbiosis in HIV-1 infected subjects. Such dysbiosis does not display obvious HIV-specific features; instead, it shares many similarities with other diseases featuring gut inflammation.
Balances: a New Perspective for Microbiome Analysis
AUTHORS: Rivera-Pinto J, Egozcue JJ, Pawlowsky-Glahn V, Paredes R, Noguera-Julian M, Calle ML.
ABSTRACT:
High-throughput sequencing technologies have revolutionized microbiome research by allowing the relative quantification of microbiome composition and function in different environments. In this work we focus on the identification of microbial signatures, groups of microbial taxa that are predictive of a phenotype of interest. We do this by acknowledging the compositional nature of the microbiome and the fact that it carries relative information. Thus, instead of defining a microbial signature as a linear combination in real space corresponding to the abundances of a group of taxa, we consider microbial signatures given by the geometric means of data from two groups of taxa whose relative abundances, or balance, are associated with the response variable of interest. In this work we present selbal, a greedy stepwise algorithm for selection of balances or microbial signatures that preserves the principles of compositional data analysis. We illustrate the algorithm with 16S rRNA abundance data from a Crohn’s microbiome study and an HIV microbiome study. IMPORTANCE We propose a new algorithm for the identification of microbial signatures. These microbial signatures can be used for diagnosis, prognosis, or prediction of therapeutic response based on an individual’s specific microbiota.
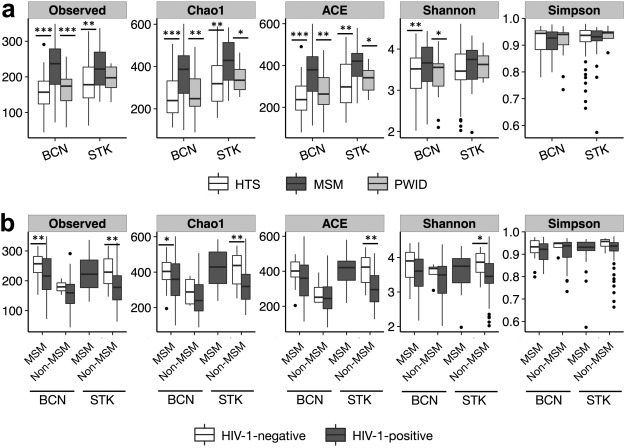
Gut Microbiota Linked to Sexual Preference and HIV Infection
AUTHORS: Noguera-Julian M, Rocafort M, Guillén Y, Rivera J, Casadellà M, Nowak P, Hildebrand F, Zeller G, Parera M, Bellido R, Rodríguez C, Carrillo J, Mothe B, Coll J, Bravo I, Estany C, Herrero C, Saz J, Sirera G, Torrela A, Navarro J, Crespo M, Brander C, Negredo E, Blanco J, Guarner F, Calle ML, Bork P, Sönnerborg A, Clotet B, Paredes R.
ABSTRACT:
The precise effects of HIV-1 on the gut microbiome are unclear. Initial cross-sectional studies provided contradictory associations between microbial richness and HIV serostatus and suggested shifts from Bacteroides to Prevotella predominance following HIV-1 infection, which have not been found in animal models or in studies matched for HIV-1 transmission groups. In two independent cohorts of HIV-1-infected subjects and HIV-1-negative controls in Barcelona (n = 156) and Stockholm (n = 84), men who have sex with men (MSM) predominantly belonged to the Prevotella-rich enterotype whereas most non-MSM subjects were enriched in Bacteroides, independently of HIV-1 status, and with only a limited contribution of diet effects. Moreover, MSM had a significantly richer and more diverse fecal microbiota than non-MSM individuals. After stratifying for sexual orientation, there was no solid evidence of an HIV-specific dysbiosis. However, HIV-1 infection remained consistently associated with reduced bacterial richness, the lowest bacterial richness being observed in subjects with a virological-immune discordant response to antiretroviral therapy. Our findings indicate that HIV gut microbiome studies must control for HIV risk factors and suggest interventions on gut bacterial richness as possible novel avenues to improve HIV-1-associated immune dysfunction.
RESOURCES